Advancing Personalized Medicine using Multiscale, Computational Modeling and Machine Learning to Determine How Biological Sex affects Respiratory Infection Outcomes
Sex hormones are critical regulators of the immune response, and estradiol levels have been linked to influenza disease outcome in males and females. My research focuses on identifying sex-specific pathways of sex hormone regulation of the immune response to infection by applying machine learning to time-series gene expression data during infection. The results of this work will enable precision medicine approaches to be developed for treating severe viral respiratory infections via hormone profile-specific drug targets. The focus on hormone profiles in the analysis enables treatments to be personalized for each individual, and beyond male and female distinctions these results can be applied to transgender individuals and pregnant persons who have unique hormone profiles. The importance of this research lies not just in using machine learning to identify new drug targets or precision medicine approaches, but also the equity and justice in developing medicine for marginalized groups who are commonly left out of medical research.
Figure 1. Disease severity in human males and females at various ages. Reproduced from Klein and Flanagan, 2016.
Figure 2. Hypothetical representation of disease progression with different interferon induction times and types. a) Early and robust interferon induction results in mild disease; b) Delayed interferon induction results in severe disease; c) Absence of interferon induction results in chronic infection; d) Early treatment with interferon therapies results in mild disease. From Gao et al, 2021.
Figure 3. Female mice treated with E2 maintained higher body weights and had higher survival than control group female mice (Sham) and female mice that were ovariectomized (Gdx). Viral titers were unchanged between the three groups, while inflammatory cytokines are significantly lower in the E2 treated group. From Robinson et al, 2011.
Sex differences in disease severity are associated with dysregulated immune responses controlled by sex hormones.
In humans of reproductive age, females experience more severe disease during influenza infection than males (Figure 1). At this same time during development, sex hormones in both sexes surge dramatically. Between males and females virus titers are unchanged at various timepoints during infection, which indicates that the immune response is the cause of severe disease. This is commonly referred to as a "cytokine storm" during which proinflammatory cytokines are overexpressed and cause excessive damage to the lung tissue, and is heavily dependent on interferon response at the site of infection (Figure 2).
Sex differences are seen in mice, where female mice have increased morbidities and mortality during influenza infection compared to age-matched males. Sex hormones have been implicated as a major regulator of the immune system, and it has been demonstrated in both mice and human cells that estradiol (E2) dramatically reduces the severity of the immune response when administered exogenously prior to or during infection (Figure 3).
The first Aim of my research is to use mice data from Robinson et al., 2011 (Figure 3) and the R21 funded research to develop an immune model capable of reproducing sex differences in disease progression. By analyzing parameter estimates and sensitivities after fitting the models to male and female data, I will be able to identify the mechanism(s) most responsible for the outcome of disease, which would be a first step in identifying novel drug targets for treating influenza infection. Additionally, I will be able to test the impact of theoretical drug targets with this model.
A limitation of mechanistic models is that they require prior knowledge about how the components in the system interact. I intend to use machine learning on the data being collected by Dr. Amie Eisfeld which will include multiple cytokines, chemokines, virus titers, and additional time-course information during influenza infection. I will train a multilayer perceptron neural network to the data to determine new sex-specific host factors of disease severity. These factors will reveal novel immune mechanisms with the potential to become novel drug targets. By utilizing machine learning algorithms I will be able to identify the best predictors of severe disease, which could also inform the structure of my mechanistic model to better simulate infection dynamics.
The pathways through which sex hormones control the immune response are likely sex- and hormone-dependent: drug targets identified would lead to the development of novel, precision medicine drugs prescribed based on patient hormone profile.
Peretz et al. demonstrated that female-derived, but not male-derived, cells responded to E2 treatment to limit influenza virus replication (Figure 4). This suggests that there exist host replication factors specific to women that could be suitable drug targets. With my experimental collaborator, Dr. John Alcorn, I will treat male and female-derived human lung epithelial cells and lung macrophages with E2 during influenza infection to generate time-series gene expression data and analyze data to identify novel host factors of influenza virus replication. The gene expression data will be analyzed in two ways: first, the gene expression data will be combined with known protein-gene interaction data from the STRING and Regulatory Circuit databases and analyzed using ProTINA software. ProTINA applies linear regression to develop scores for all the protein-gene interactions in the model, with the highest scores suggesting the most likely protein/gene affecting the experiment phenotype. Applied to the data here, ProTINA can identify the most important host factors regulating virus replication. However, network-based tools like ProTINA are limited by our knowledge of protein-gene interactions.
In our second approach, the gene expression data, virus growth rates, and experimental conditions will be analyzed with a machine learner to predict virus growth rate for each patient/donor. I will use a multilayer perceptron neural network, but will also consider other algorithms, such as Support Vector Machines (SVMs). I hope to reveal distinct proteins, or distinct immune pathways, that cause enhanced virus replication between males and females, which can be used to develop personalized treatments based on sex or even hormone profile.
Though initially this work will focus on cisgender males and females, treatments based on patient hormone profile analyses can be given to transgender persons. While there is a significant knowledge gap surrounding female health, this gap is even greater for transgender persons. The knowledge generated from this project can be utilized in multiple fields, including drug discovery, drug repurposing, immunology, and precision medicine. There is no doubt that new treatments are needed for viral diseases, and any key pathways and drug targets identified as part of this work would certainly influence drug discovery projects. Identifying different treatment targets or strategies based on sex or hormone profile is a first step towards precision medicine, and this research will help turn precision medicine into a reality in the clinical setting. There are numerous disease that disproportionally affect a particular sex (Figure 5), so this work could be broadly applied to find sex-specific treatments and identify novel, sex-specific immune pathways.
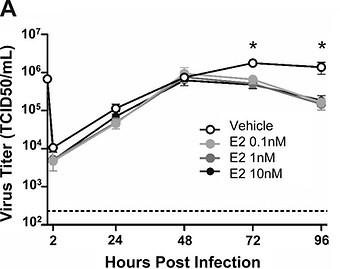
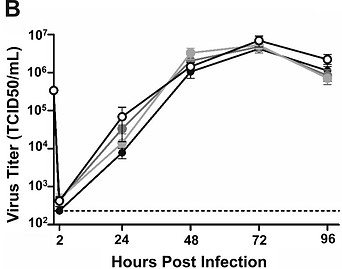
Figure 4. Female, not male, human nasal epithelial cells (hNECs) respond to E2 treatment and result in decreased viral titer during influenza infection. All E2 concentrations tested resulted in decreased viral titer for the female cells, compared to untreated cells. From Peretz et al, 2016.
Figure 5. Multiple types of diseases and disorders have a sex bias. The majority of these diseases have not been investigated in a sex-specific manner, leaving a significant knowledge and data gap that negatively impacts progress in treatment. From Mauvais-Jarvis et al, 2020.